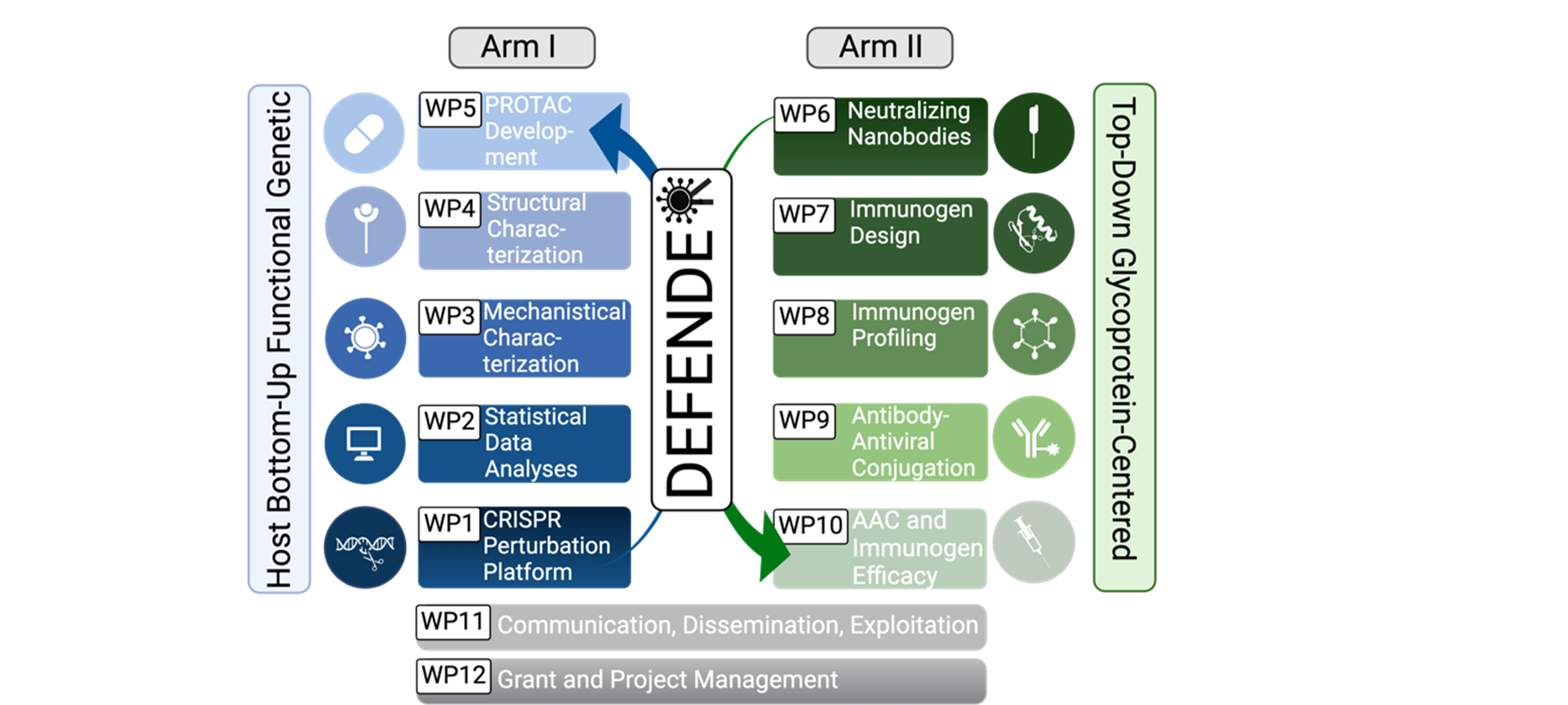
DEFENDER’s two-armed platform is being applied to (re-)emerging viruses, integrating host- and virus-centered strategies to reveal novel interaction for new antivirals and immunogens.
Partner roles and contributions
Adriano Aguzzi (UZH)
Leads WP1 (“Arrayed CRISPR perturbation platform for the identification of candidate host cell entry factors for emerging viruses”). His objectives are to standardize DEFENDER’s high-throughput arrayed CRISPR platform and identify host factors influencing virus entry and uptake. With extensive expertise in CRISPR perturbation assays, his group also contributes to WP3.
Lars Kaderali (UMG)
Leads WP2 (“Statistical data analysis, bioinformatics and machine learning approaches for putative drug target identification”). His work involves data normalization and statistical analysis of CRISPR perturbation data, identifying host factors mediating virus entry (both broad and virus-specific), and developing predictive models for rapid drug target identification in emerging viruses.
Stephanie Pfänder (LIV)
Leads WP3 (“Mechanistic characterization of host factors impacting virus entry in vitro”), including studies in immortalized and authentic systems, as well as combinatorial gene analyses. LIV also coordinates WP11 (“Communication, dissemination, and exploitation”) and WP12 (“Grant and project management”). The Pfänder lab contributes to WPs 1, 2, 4, and 5, focusing on ZIKV.
Petr Chlanda (UKHD)
Leads WP4 (“Structural and topological characterization of cellular targets using cellular cryo-electron tomography (cryo-ET)”), which includes generating fluorescent reporter viruses for cryo-CLEM approaches, applying cryo-CLEM and cryo-ET to study molecular mechanisms inside infected cells, and mapping the subcellular localization of identified targets. UKHD also contributes to WPs 6 and 9.
Mark Brönstrup (HZI)
Leads WP5 (“PROTAC development and testing as innovative antiviral strategies”), focusing on the development of host-targeting PROTACs, their antiviral activity testing, and safety/potency studies in animal models. HZI also coordinates WP9 (“Antibody–antiviral conjugation”), which involves selecting epitopes, preparing nanobodies and AACs, using cryo-ET to study inhibitory mechanisms, and developing one monoclonal AAC for preclinical testing.
Marie Flamand (IP)
An expert in DENV infection, the Flamand lab contributes to WPs 1–5.
Christine Goffinet (LSTM)
Specialized in CHIKV, the Goffinet lab contributes to WPs 1–5.
Eike Steinmann (RUB)
With expertise in molecular virology, molecular biology, and reverse genetics (YFV), the Steinmann lab contributes to WPs 1–5.
Thomas Krey (UZL)
Leads WP6 (“Identification of broadly neutralizing nanobodies”), including AI-based design and characterization of “headless” glycoproteins, expression of stabilized viral glycoproteins, functional studies of nanobodies/antibodies, and cryo-EM mapping of key interactions.
Also leads WP8 (“Profiling of designed immunogens”), covering immunogen multimerization, biochemical/structural characterization, and immunological profiling. His lab additionally contributes to WPs 7, 9, 10, and 12.
Thomas Strecker (UMR)
Specialized in LASV and other highly pathogenic viruses, the Strecker lab contributes expertise in BSL-4 operations, mechanisms of LASV infection, and therapeutic antibody development in WPs 1–6, and 9.
Bruno Correia (EPFL)
Leads WP7 (“Immunogen design”), focusing on computational design of immunogens with enhanced antigenic properties, mid-throughput screening and biochemical characterization, as well as antigenic profiling and structural studies. His expertise in computational protein engineering also supports WPs 6, 8, 9, and 10.
Andrea Maisner (UMR)
As an expert in studying the cell biology of NiV in vitro, the Maisner lab has expertise in BSL-4 infections and contributes to WPs 1–6, and 9, investigating host factors that influence NiV replication and testing the antiviral activity of PROTACs, nanobodies, and AACs in cell culture.
Alexandra Kupke (UMR)
Specialized in in vivostudies of highly pathogenic viruses, the Kupke lab contributes to WPs 5, 8 and 10 by evaluating antiviral activities of PROTACs, AACs and nanobodies against NiV.
Lisa Oestereich (BNI)
Leads WP10 (“AAC and immunogen efficacy in vivo”), evaluating therapeutic effects of AACs and protective effects of immunogens against NiV and LASV. The Oestereich lab, specialized in innate immune responses and animal modeling of LASV, provides expertise in in vivo models and assays.
All partners are actively involved in every work package (WP), including WP11 (Communication, Dissemination, and Exploitation) and WP12 (Management), ensuring effective collaboration, research harmonization, synergies, and informed decision-making.